Home
Research Interests
We are a multidisciplinary research group based in the School of Physics and Astronomy and motivated to understand the physics of life. We are developing tools to explore multiscale scale mechanics, single molecule manipulation techniques and neutron scattering to explore the physics of living systems. These powerful techniques are used to study hierarchical biomechanics, self-assembly and the structure and dynamics of molecules in aqueous solutions, in both simple and complex systems. We are members of the Astbury Centre for Structural Molecular Biology and the Bragg Centre for Materials Research. We gratefully acknowledge funding from the EPSRC, STFC, BBSRC and ERC. Read about our latest paper on Hierarchical biomechanics Here!
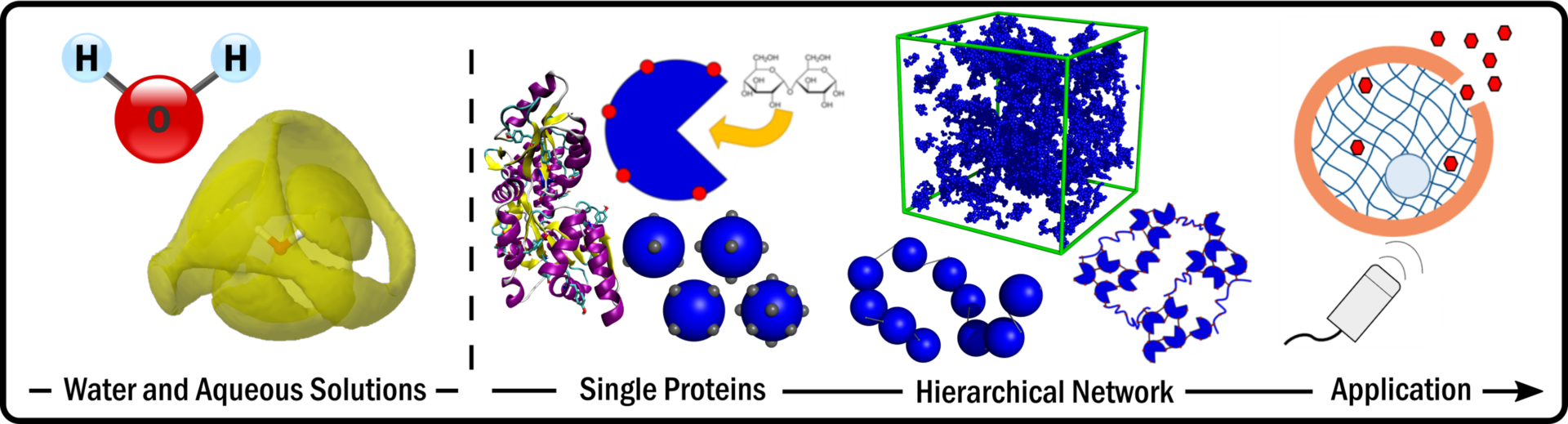

Research projects
- Engineering protein-based hydrogels
- Water structure
- Life in extreme environments
- Single molecule force spectroscopy
- Cropreservation
- Biomolecule self-assembly
More about our Research >
Contact details
- L.Dougan@leeds.ac.uk
- +44 (0) 113 343 8958
- School of Physics and Astronomy
Bragg Building
University of Leeds
Leeds LS2 9JT
UK
